Synthetic DNA sequences
2018-

An important and largely unsolved problem in synthetic Biology is how to control gene expression levels by cellular programming. Endogenously, such control is achieved by way of regulatory elements such as promoters, enhancers and insulators. One of the main hallmarks of such elements is their accessibility to factors promoting transcription of cognate genes. To achieve cellular programming through the manipulation of regulatory elements, it is therefore desirable to dictate the accessibility patterns of elements across cellular conditions of interest.
We propose a fully data-driven framework for designing synthetic human sequence elements with predicted cell-context specific chromatin accessibility behavior. We apply a comprehensively annotated high-resolution map of accessible genomic elements as the basis for a training set to learn and apply the foundational rules regarding how regulatory elements are encoded in the human genome. This approach results in highly variable pools of synthetic sequences, not observed in nature.
We use a supervised adaptation strategy to evolve these sequences towards multiple pre-defined cellular contexts. We show that this tuning process not only retains characteristics of endogenous regulatory sequences, but often strengthens them through an increased abundance of relevant regulatory signals.
Our approach and its current results provide a foundation for subsequent experimental validation through the targeted integration of synthetic sequences in genomic loci in relevant cellular conditions.
Work with Peter Bromley
Material available for download
Code base
All code is available on GitHub
Curated chromatin accessibility datasets
For this project we make use of a dataset of chromatin accessibility obtained across 733 human biosamples, containing more than 3.5 million DNase Hypersensitive Sites (DHSs). These data were previously published (paper) and are available here.
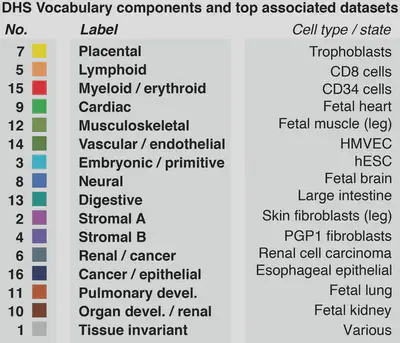
An important aspect of these data is an annotation of accessibile elements in terms of 16 components, each corresponding to a different cellular context (e.g. ‘cardiac’, ’neural’, ’lymphoid’):
To aid in efforts towards understanding and generating regulatory sequences, we curated a subset of these data and provide it to the wider community. This subset consists of fixed-length 200bp sequences selected from the total collection of 3.5M+ sequences based on their accessibility confidence level and relative specificity for individual DHS components.
Specifically, we have the following datasets available:
- training set: 160k sequences, 10k per NMF component (chr3-chrY, .csv.gz)
- validation set: 16k sequences, 1k per NMF component (chr2 only, .csv.gz)
- test set: 16k sequences, 1k per NMF component (chr1 only) .csv.gz)
Each of these contains the genomic locations (human genome assembly hg38, first 3 columns) of accessible genome elements,
their majority NMF component (column: component, see legend figure above) as well as their 200bp nucleotide sequence (column: raw_sequence).
| Column | Example | Description |
|---|---|---|
seqname |
chr16 | Chromosome |
start |
68843660 | Start position |
end |
68843880 | End position |
DHS_width |
220 | Width of original DHS |
summit |
68843790 | Position of center of mass of DHS |
total_signal |
122.770678 | Sum of DNase-seq signal across biosamples |
numsamples |
61 | Number of biosamples with the DHS |
raw_sequence |
GAGGCATTG… | 200bp extracted nucleotide sequence |
component |
1 | Dominant DHS Vocabulary component |
proportion |
0.767371514 | Proportion of NMF loadings in this component |
Please refer to the DHS Index project page for more information and details on the full dataset.