Annotated regulatory Index
2017-2020
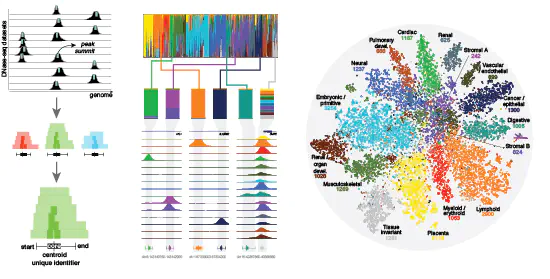
DNase I hypersensitive sites (DHSs) are generic markers of regulatory DNA and contain genetic variations associated with diseases and phenotypic traits. We created high-resolution maps of DHSs from 733 human biosamples encompassing 438 cell and tissue types and states, and integrated these to delineate and numerically index approximately 3.6 million DHSs within the human genome sequence, providing a common coordinate system for regulatory DNA.
Here we show that these maps highly resolve the cis-regulatory compartment of the human genome, which encodes unexpectedly diverse cell- and tissue-selective regulatory programs at very high density. These programs can be captured comprehensively by a simple vocabulary that enables the assignment to each DHS of a regulatory barcode that encapsulates its tissue manifestations, and global annotation of protein-coding and non-coding RNA genes in a manner orthogonal to gene expression.
Finally, we show that sharply resolved DHSs markedly enhance the genetic association and heritability signals of diseases and traits. Rather than being confined to a small number of distal elements or promoters, we find that genetic signals converge on congruently regulated sets of DHSs that decorate entire gene bodies. Together, our results create a universal, extensible coordinate system and vocabulary for human regulatory DNA marked by DHSs, and provide a new global perspective on the architecture of human gene regulation.
Meuleman et al., Nature (2020)
Work with Sasha Muratov, John Stamatoyannopoulos and others.
Material available for download
-
DHS Index and Vocabulary of ~3.6M DHSs (tsv, hg38)
Column Example Description seqnamechr1 Chromosome start1782520 Start position end1782770 End position identifier1.10643 Unique identifier (chr#.position%) mean_signal1.030869481 Mean DNase-seq signal per biosample (“confidence score”) numsamples54 Number of biosamples with a DHS summit1782650 Estimated DHS summit position core_start1782590 Start position of core-region core_end1782710 End position of core-region componentDigestive Main DHS Vocabulary component -
Data browsers (Altius browser, UCSC browser tracks)
-
733 biosample metadata (html, pdf, tsv, xlsx, Google spreadsheet)
-
DHS-by-biosample matrices
Rows correspond to DHS Index elements, columns correspond to rows in biosample metadata (both in order).- Presence/absence matrix (R sparse Matrix format, Matrix Market format, raw text format)
- Signal matrix (R sparse Matrix format, raw text format)
-
Per-biosample and per-component data
- DHS Index elements found in specific biosamples:
Column `biosample_signal` reports the normalized DNase-seq signal for the individual biosample.
- DHS Index elements annotated with specific DHS components:
By default, each DHS is annotated with a single (dominant) component only. DHSs may occur in biosamples of other components too! Refer to the data matrices listed above for the full picture.
- DHS Index elements found in specific biosamples: